Translate this page into:
The incidence of TP53 mutations in patients with chronic lymphocytic leukemia in Iran
*Corresponding author: Behzad Poopak, Department of Hematology, Islamic Azad University, Tehran University of Medical Sciences, Tehran, Iran. bpoopak@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Bayati N, Poopak B, Emtiazi N, Ghiass MA. The incidence of TP53 mutations in patients with chronic lymphocytic leukemia in Iran. Asian J Oncol. 2024;10:2. doi: 10.25259/ASJO_33_2023
Abstract
Objectives
Chronic lymphocytic leukemia (CLL) is linked to a highly variable disease course regarding responses to chemoimmunotherapy and clinical outcomes. Mutations in TP53 and/or deletions in chromosome 17p locus [del(17p)] may lead to loss of a TP53 allele. We investigated TP53 mutations in patients with CLL.
Material and Methods
Thirty patients with CLL, aged 40–84 years, were included. Immunophenotyping of B cells was done using flow cytometry by CD5, CD10, CD19, CD20, CD23, CD49d, CD38, FMC7, and CD200 markers. Bone marrow aspiration and peripheral blood smear examination were also performed. DNA was extracted and sequencing was done using the Sanger method. Structural aberrations were investigated using the FISH method.
Results
TP53 mutations had a frequency of eight cases (23.3%), three of which were missense mutations (37.5%), three were intronic mutations (37.5%), and two were silent mutations (25%). Peripheral B lymphocytes had a mean of 82% with 4.59% prolymphocytes.
Conclusion
Accurate testing for TP53 mutations [TP53 and del(17p) mutations] before every treatment line enables us to make proper therapeutic decisions to improve patient outcomes.
Keywords
Chronic lymphocytic leukemia
TP53 mutation
Deletion
Sanger sequencing
INTRODUCTION
Chronic lymphocytic leukemia (CLL) disease has an extremely heterogeneous nature. Although some cases survive for over ten years with no treatment, some experience poor outcomes and rapid disease progression in spite of successful chemoimmunotherapy.[1,2] The reason for this heterogeneity is different genetic aberrations recognized in patients with CLL. It can be found in old population and it is rare in cases with age >40 years old.[3,4] Deletions in chromosome 17p [del(17p)] cause the loss of the TP53 gene, encoding the tumor-suppressor protein p53, which is linked to a poor prognosis.[5] Also, TP53 mutations are involved in poor prognosis regardless of the existence of del(17p). In general, such mutations and deletions are illustrated in TP53 as defective.[6]
TP53, also known as tumor protein 53 or p53, is a crucial tumor suppressor gene that plays a fundamental role in preventing the development and progression of cancer. This gene is located on chromosome 17p13.1 and encodes the p53 protein, which acts as a transcription factor to regulate the expression of numerous genes involved in cell cycle control, DNA repair, apoptosis, and genomic stability.[7] TP53 mutations are considered one of the strongest predictive and prognostic markers to guide treatment lines in CLL and are linked to a decrease in survival and resistance against chemoimmunotherapy. Numerous studies have demonstrated the TP53 mutation analysis clinical value in CLL. Recently, the treatments that are efficient in patients harboring TP53 mutations include the B Cell Lymphoma 2 (BCL2) inhibitor venetoclax, ibrutinib as Bruton tyrosine kinase (BTK) inhibitor, and idelalisib as the phosphatidylinositol 3-kinase (PI3K) inhibitor. Therefore, recognizing TP53 mutations is crucial to determine the best treatment line and decision for patients.[8,9] There are different diagnostic methods that are used routinely like Sanger sequencing and next-generation sequencing, which also use fluorescence in situ hybridization (FISH) to identify del(17p), but a proportion of patients (30–40%) harbor TP53 aberration in the absence of del(17p). Thus, in addition to evaluating del(17p), we need to assess TP53 mutations using Sanger sequencing to select the appropriate treatment for patients.[10,11] In this study, we aimed to evaluate the incidence of TP53 mutations and lymphocyte counts in patients with CLL in a small Iranian population.
MATERIAL AND METHODS
This cross-sectional study was performed in the Peyvand clinical laboratory from Jan 2018 to Feb 2019. Thirty patients at the diagnosis stage (aged 40–84 years) were included in the present study. Following the WHO guidelines, CLL was diagnosed by blood counts and smears. Three patients had bone marrow samples and 27 patients had peripheral blood samples, which had a minimum sample volume of 2 cc and were collected in vials containing the K2-ethylenediaminetetraacetic acid (EDTA) anticoagulant. This study was approved by the Ethics Committee of Tarbiat Modares University (IR.MODARES.REC.1397.227) and all patients signed written informed consent prior to sample collection. This study was done according to the ethical standards of the relevant national and institutional guidelines on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
Flow cytometry
The samples were stained with 20 µL of each monoclonal antibody for 15 minutes at 37°C and 15 minutes at room temperature, then erythrocytes were lysed by adding 1.1 µL of fluorescence-activated cell sorter lysing solution (Becton Dickinson, Lincoln Park, NJ, USA). After staining, the cells were washed three times in phosphate-buffered saline (PBS)[12] and immediately analyzed using Becton Dickinson (BD) Fluorescence Activated Cell Sorting (FACS) Lyric™ Flow cytometry from Beckman Colter (USA) [Figures 1a–1b]. Immunophenotyping of B lymphocytes was done using CD5, CD10, CD19, CD20, CD23, CD38, CD49d, CD200, and FMC7 markers. Complete cell blood counts were assessed by Mindray-6800 differential hematology analyzer. We also conducted bone marrow aspiration and peripheral blood smear examination (if available).
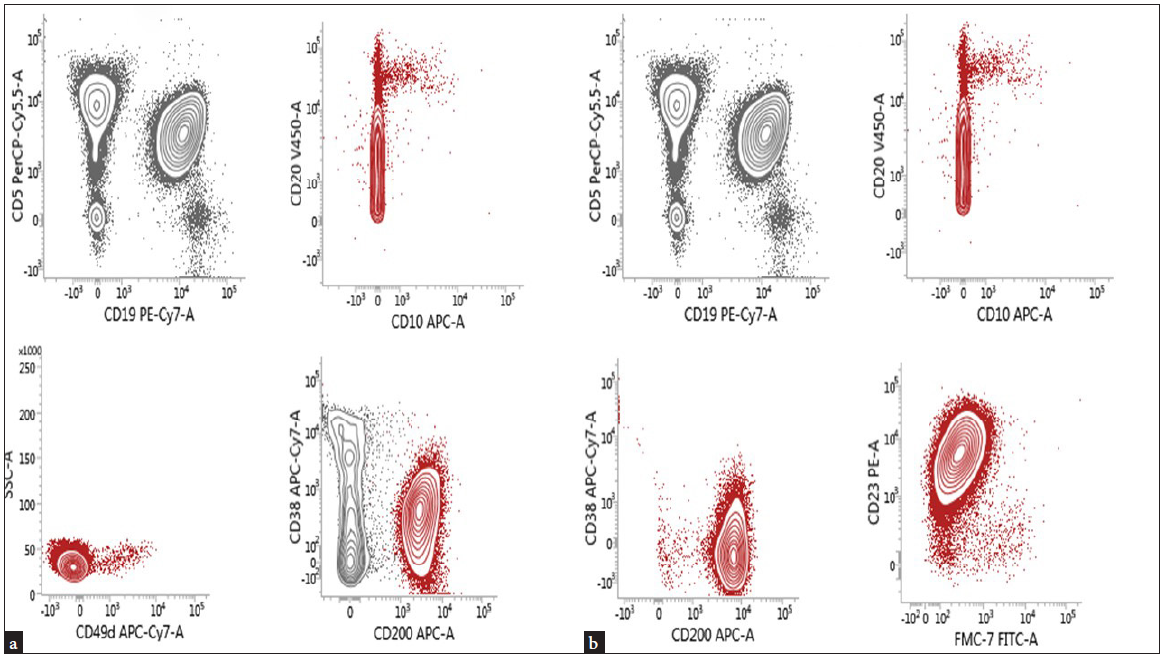
- Flow cytometry markers for B-cell counts.
DNA extraction and sequencing
DNA was extracted using the salting out method. After extraction, quantity of DNA was assessed by Nanodrop (Optical Density (OD) 260/280) and 1.7<OD<2.2 was used for Polymerase Chair Reaction (PCR) test. Primer sequences [Table 1] and the protocol to perform PCR are present on the International Agency for Research on Cancer (IARC) TP53 site (http://p53.iarc.fr/ProtocolsAndTools.aspx). The PCR protocol was as follows: The first step involves the initial denaturation of the DNA template at 95°C for 5 min and 95°C for 40 sec. The second step is the annealing of the primers to the targeted DNA sequence on the template at 66°C for 40 sec (Note: The annealing temperature is routinely 5°C below the primer Tm). The third step is the extension of the template sequence by Taq DNA polymerase at 72°C for 40 cycles.
| Amplification Region | Primer Pairs Direction | Lengths |
|---|---|---|
| Exon 6 |
F: 5’-tgttcacttgtgccctgact-3’ R: 5’-ttaacccctcctcccagaga-3’ |
467 bp |
| Exon 7 |
F: 5’-cttgccacaggtctccccaa-3’ R: 5’-aggggtcagaggcaagcaga-3’ |
237 bp |
| Exon 7 |
F: 5’-aggcgcactggcctcatctc-3’ R: 5’-tgtgcagggtggcaagtggc-3’ |
177 bp |
| Exon 8 |
F: 5’-ttccttactgcctcttgctt-3’ R: 5’-aggcataactgcacccttgg-3’ |
231 bp |
Bidirectional (Forward+Reverse) sequencing analysis was assigned and the chromatograms produced by Sanger sequencing were assessed. Sanger sequencing data were analyzed by GLASS (ERIC TP53 Network provides). Structural aberrations were investigated by the FISH method.
Statistical analyses
Statistical analyses were carried out with Statistical Package for the Social Sciences (SPSS) version 23.0 (SPSS Inc., Chicago, Illinois, USA). The normality of the data was evaluated by the Kolmogorov–Smirnov test. The Kruskal–Wallis test was used to assess significant intergroup variations. The Fisher’s exact test was used for between-group mutations.[13] The Pearson test was used to determine correlations between groups. A P-value of less than 0.05 was considered statistically significant.
RESULTS
Eighteen males and 12 females were evaluated with an age range of 40–84 years [Table 2].
| N:30 | ||
|---|---|---|
| Age | Mean | 40–84 |
| Sex | Female | 12 (44%) |
| Male | 18 (56%) | |
| Staging | 0 | 13 (43%) |
| I | 6 (20%) | |
| II | 1 (4%) | |
| III | 4 (13%) | |
| IV | 6 (20%) |
Leukocytosis in samples ranged from 12.35 to 559.9 × 103 cells/µL (The median was 114.0 × 103 cells/µL). Hb ranged from 6.8 to 16.0 g/dL with an average of 11.9 g/dl. Platelet (PLT) count ranged from 27 to 307 × 103 cells/µL with a mean of 188 × 103 cells/µL. Peripheral blood lymphocytes were detected with a mean of 82% and 4.59% prolymphocytes [Table 3].
| Parameters | Mean |
|---|---|
| TLC (× 103 cells/µL) | 114.0 |
| Hb (g/dl) | 11.9 |
| PLT (× 103 cells/µL) | 188.0 |
| PB lymph. % | 82.0 |
| Pro lymph. % | 4.59 |
| Atypical lymph. % | 1.0 |
TLC: Total leukocyte count, PLT: platelet, PB: peripheral blood, Hb: hemoglobin, lymph.: lymphocyte, CLL: Chronic lymphocytic leukemia.
Seven cases (23.3%) were positive for the TP53 gene mutation, two of which were missense mutations (28.5%) [Figure 2a], two were silent mutations [Figure 2b], and three were intronic mutations (43%) (28.5%) [Figure 2c]. All mutations were clustered within exons 6–8 that encode the DNA-binding domain [Table 4, Figure 3].
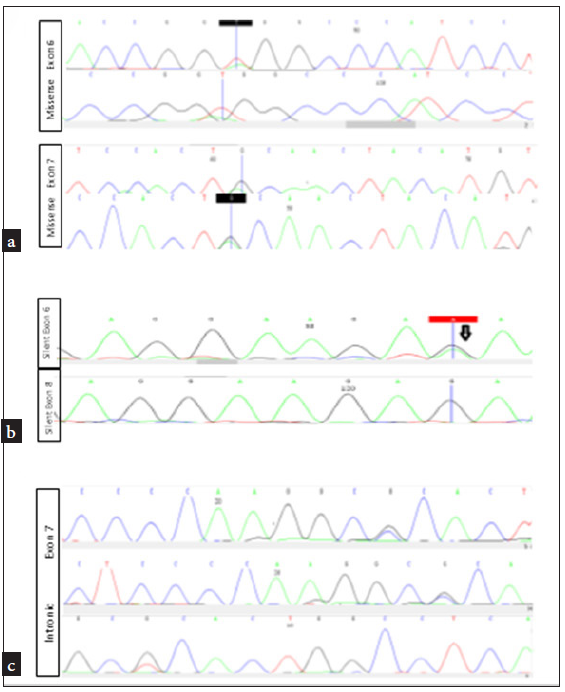
- (a) The TP53 gene mutation was positive in seven cases, two of which were missense mutations (28.5%), (b) two were silent mutations (43%), (c) and three were intronic mutations 28.5%.
| Exon | TP53 mutations | ||||
|---|---|---|---|---|---|
| Missense | Substitution | Silent | Substitution | Intronic | |
| Exon 6 | 1 | p.(I195T) | 1 | p.(R213R) | - |
| Exon 7 | 1 |
p.(R249W) p.(Y234C) |
- | 3 | |
| Exon 8 | - | - | 1 | p.(E287E) | - |
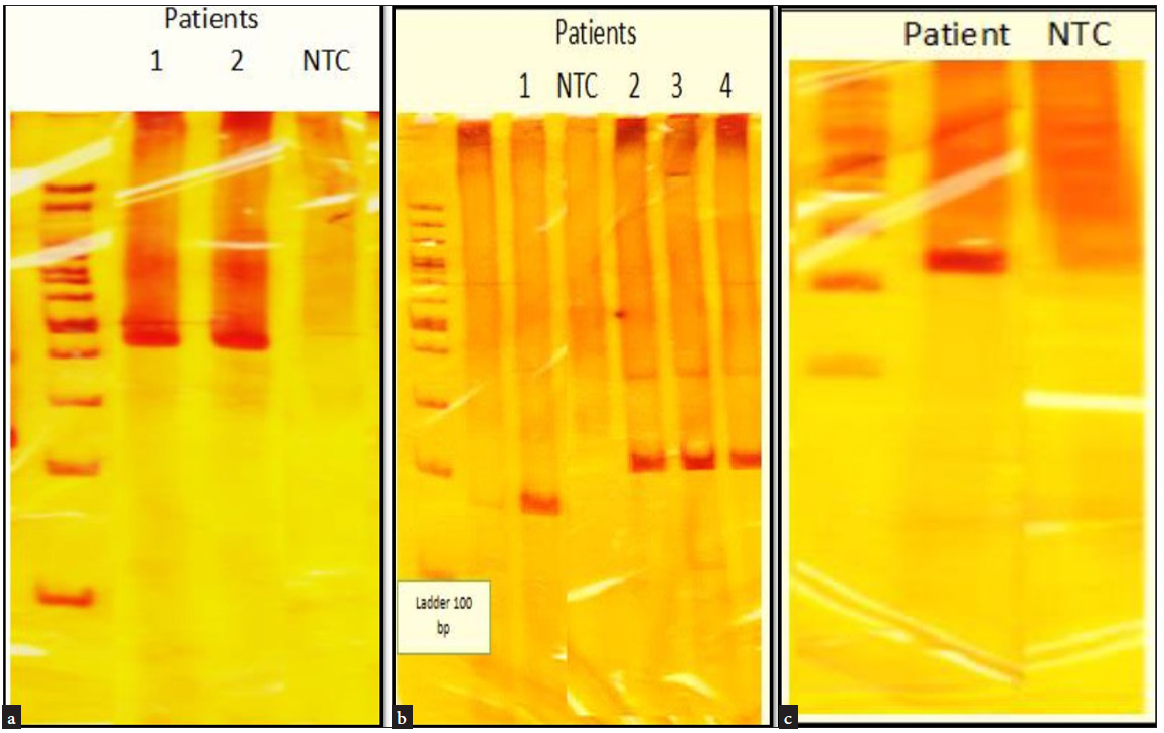
- (a) PCR gel of two patients with mutation in exon 6 with product length of 467 bp. (b) Three patients had a mutation in exon 7 with a primer with product length of 237 bp, and one patient had a mutation in exon 7 with product length of 177 bp. (c) PCR gel of one patient with mutation in exon 8 with product length of 231 bp, NTC: Negative control, PCR: Polymerase Chain reaction, bp: Base pair.
Five (62.5%) mutations were detected in male patients and three (37.5%) were detected in female patients. Although hematologic parameters such as leukocyte increased, hemoglobin (Hb) and PLT decreased in patients with mutations in comparison to other patients. Besides, an increase was seen in prolymphocytes, but no significant association was detected between TP53 gene mutation and age, gender, and hematologic parameters. Most of the patients with the mutation were in the progressive stage of the disease according to Rai stage classification (III and IV) [Table 5].
| TP53 Mutation | P-value | Sig. | |||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Age (years) | 63 | 65 | 0.670 | NS | |
| Sex | Female | 3 (37.5%) | 9 (41 %) | 0.427 | NS |
| Male | 5 (62.5%) | 13 (59%) | NS | ||
| WBC (× 103 cell/µL) | 98.41 | 92.78 | 0.725 | NS | |
| HB (g/dl) | 10.8 | 12.3 | 0.241 | NS | |
| PLT (× 103 cell/µL) | 154 | 193 | 0.434 | NS | |
| Lymphocyte (%) | 71.7 | 85.6 | 0.051 | NS | |
| Pro Lymphocyte (%) | 8.5 | 7.1 | 0.434 | NS | |
P-value <0.05: Significant (S), P-value >0.05: Non-significant (NS), WBC: White blood cells, PLT: platelet, HB: hemoglobin.
We detected del(17p) structural aberration using FISH in three patients and detected del(17p) in all of them. In one patient, both TP53 mutations and del(17p) were documented, while del(17p) was found only in the two remaining patients. According to Sanger sequencing data, about 80% of those with del(17p) carry TP53 mutation as well. Some patients who had prolymphocyte counts >40% showed the following mutations: One had only del(17p) and another one had TP53 mutation. Some patients with del (17p) and TP53 mutations have been shown high aberrant expression of immunophenotyping markers, such as CD38 and CD49d.
DISCUSSION
The results of this study regarding the TP53 gene in 30 patients with CLL indicated that there was a mutation in 23.3% of the patients. Also, in a study by Wu and his colleagues in Taiwan in 2017, they reported the percentage of mutation in 83 patients to be 20.5%.[14] Also, in a study published by Stengel et al. in 2016, the percentage of mutations in 332 patients was reported as 10%.[15] Also, 23.3% of patients were positive for TP53 gene mutation (seven patients), which is higher than in the Western community. Rossi et al.[16] declared TP53 mutations in merely 10% of cases at diagnosis, which can indicate a difference in disease biology. FISH is highly used to diagnose chromosomal abnormalities, like 17p deletion in CLL; however, no complete overlap is available between del(17p) and TP53 mutation, approximately 30–40% of cases harbored TP53 mutation without del(17p). In addition, TP53 mutations are linked to a poor prognosis independent of the existence of del(17p), thus a sequencing method is necessary to examine TP53 mutations’ status. With a sensitivity of approximately 15%–20% of mutated DNA, direct sequencing can be regarded as the standard approach to detect TP53 mutations. We identified three patients with del(17p) and one patient with TP53 mutation. Also, these patients were in advanced stages of CLL. Collectively, del(17p) is the commonest genetic abnormality affecting the TP53 gene in CLL, which accounts for about two-thirds of the cases.[6,17]
Rigolin et al.[18] reported a strong association between advanced CLL stages and aberrant karyotyping, the need for treatment, and also shorter overall survival and worse prognosis. All mutations were clustered within exons 6–8 that encode the DNA-binding domain. Also, substitutions were in the gene’s coding region. Missense mutations in the TP53 coding region are known as the most common mutations, which change one amino acid in the P53 protein. Such mutations induce the expression of P53 protein that is not able to obtain oncogenic function and activate tumor suppressor response.[19] Six hotspot codons, including codons 175, 249, 245, 273, 248, and 282, have been reported to affect frequency. This is associated with the TP53 mutational profile in CLL.[20,21]
CD38, known as an apoptotic transmembrane glycoprotein, is expressed by normal B cells. In addition, CD38 encourages B cell proliferation and survival. This threshold predicts a poor prognosis because it predicts shorter treatment lag times and lower response rates.[22] Also, CD49d is another aberrant immunophenotyping marker that has a strong correlation with the migratory potential of CLL cells and unfavorable outcomes. CD49d expression shows a significant relationship with the Rai stage of the patients since patients positive for CD49d are in higher stages than patients negative for CD49d, so these markers are widely used as prognostic markers.[23] In our study, two patients had del(17p), and one patient had TP53 mutation at higher disease stages (III, IV), and high CD49d and CD38 expression. Furthermore, 75% of patients with TP53 mutation and del(17p) were significantly related to clinical prognostic data, like hepatomegaly, splenomegaly, and lymphadenopathy. Xia et al.[24] declared an advanced Binet stage linked to TP53 mutation.
In this study, we detected two patients with del(17p) and one patient had TP53 mutation who had increased prolymphocyte (44%–50%). David Oscier et al.[25] reported a relationship between elevated prolymphocytes and NOTCH1 mutations as a new and unexpected result. So further research may figure out an association between increased prolymphocyte and abnormal cytogenetic in patients with CLL as a prognostic factor. We consider hematologic parameters in patients with mutation and no mutation. Finally, we indicate that an increase in white blood cells (WBC) and a decrease in Hb and PLT in patients with the mutation may be more sensible than no mutation. Olfat M. Hendy et al.[17] reported a significant association between increased WBCs and decreased Hb and PLT in patients with TP53 mutation in comparison to patients with no mutation. TP53 alterations in CLL make reasonable the use of new therapeutic targets such as ibrutinib, idelalisib, acalabrutinib, and venetoclax, for targeting B cell signaling. Therefore, the treatment approach may change by detecting subclonal TP53 mutations.[26] The study may have some limitations as follows: small sample size, which can affect the generalizability of the findings. In addition, the ratio of females and males was not equal.
CONCLUSION
The incidence of TP53 mutations serves as a reliable predictor of chemorefractoriness in CLL patients. Our research, along with the analysis of publicly available CLL databases, reveals that the majority of these mutations occur in exons 6–8, with mutations at other sites being rare exceptions. It can be considered a valuable prognostic tool for CLL patients who are candidates for treatment. The clinical significance of TP53 mutations necessitates their inclusion as a routine testing procedure before initiating any line of treatment. However, it is crucial to note that our findings should be validated through larger-scale studies encompassing a wider range of TP53 mutation variants. By expanding our investigations to include other mutation variants, we can gain comprehensive insights into the clinical implications of TP53 mutations and refine our strategies for managing CLL patients. Ultimately, such advancements will contribute to improved patient outcomes and a more tailored approach to CLL treatment.
Acknowledgments
The current paper was supported by the Payvand clinical laboratory. The author thanks Behzad Poopak for his support in obtaining scientific data presented here and at Tarbiat Modares University.
Authors’ contributions
NB conceived the study, conceptualized and wrote the first manuscript, prepared tables and figures, and revised the final manuscript for important intellectual contents; NE, BP, and MAG helped in further conceptualization of the manuscript; BP and MAG supervised the study. All authors read and approved the final version of the manuscript.
Ethical approval
Institutional Review Board approval is not required.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
References
- Detection of somatic TP53 mutations and 17p deletions in patients with chronic lymphocytic leukemia: A review of the current methods. Hematol Transfus Cell Ther. 2020;42:261-8.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- TP53 gene mutation analysis in chronic lymphocytic leukemia by nanopore MinION sequencing. Diagn Pathol. 2016;11:1-9.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia—update on methodological approaches and results interpretation. Leukemia. 2018;32:1070-80.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Managing patients with TP53-deficient chronic lymphocytic leukemia. J Oncol Pract. 2017;13:371-7.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical flow-cytometric testing in chronic lymphocytic leukemia. Methods Mol Biol. 2019;2032:311-21.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- TP53 aberrations in chronic lymphocytic leukemia: an overview of the clinical implications of improved diagnostics. Haematologica. 2018;103:1956.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10:37-50.
- [CrossRef] [PubMed] [Google Scholar]
- 17P deletion and TP53 gene mutation (17P/TP53) testing behaviour and treatment patterns for chronic lymphocytic leukemia (CLL) patients in France, Germany, Italy, Spain and UK (EU5) Ann Oncol. 2016;27:vi317.
- [Google Scholar]
- 17p deletion strongly influences rituximab elimination in chronic lymphocytic leukemia. J Immunother Cancer. 2019;7:1-5.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2021;32:23-33.
- [CrossRef] [PubMed] [Google Scholar]
- ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia. 2012;26:1458-61.
- [CrossRef] [PubMed] [Google Scholar]
- Interleukin-21 receptor might be a novel therapeutic target for the treatment of rheumatoid arthritis. Journal of Experimental & Clinical Medicine. 2014;6:57-61.
- [PubMed] [Google Scholar]
- Frequency of CD4+ and CD8+ T cells in iranian chronic rhinosinusitis patients. Allergy Asthma Clin Immunol. 2018;14:1-11.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Distinct molecular genetics of chronic lymphocytic leukemia in taiwan: Clinical and pathogenetic implications. Haematologica. 2017;102:1085.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2017;31:705-11.
- [CrossRef] [PubMed] [Google Scholar]
- The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clinical cancer research. 2009;15:995-1004.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of TP53 gene mutation in chronic lymphocytic leukemia patients. The Egyptian Journal of Hospital Medicine. 2022;89:5807-12.
- [PubMed Central] [Google Scholar]
- Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia with “normal” FISH: correlations with clinicobiologic parameters. Blood. 2012;119:2310-3.
- [CrossRef] [PubMed] [Google Scholar]
- Flow cytometric expression of CD49d in newly diagnosed chronic lymphocytic leukemia and its correlation with established prognostic markers. J Lab Physicians. 2022;14:435-42.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Recommended guidelines for validation, quality control, and reporting of TP53 variants in clinical practice. Cancer Res. 2017;77:1250-60.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- TP53 mutation profile in chronic lymphocytic leukemia: evidence for a disease specific profile from a comprehensive analysis of 268 mutations. Leukemia. 2010;24:2072-9.
- [CrossRef] [PubMed] [Google Scholar]
- CD38 and chronic lymphocytic leukemia: A decade later. Blood. 2011;118:3470-8.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Association of CD49d expression with clinicopathological features of chronic lymphocytic leukemia patients in the Iranian population. Int J Physiol Pathophysiol Pharmacol. 2020;12:32.
- [PubMed] [PubMed Central] [Google Scholar]
- Frequencies of SF3B1, NOTCH1, MYD88, BIRC3 and IGHV mutations and TP53 disruptions in Chinese with chronic lymphocytic leukemia: disparities with Europeans. Oncotarget. 2015;6:5426.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The morphology of CLL revisited: the clinical significance of prolymphocytes and correlations with prognostic/molecular markers in the LRF CLL4 trial. Br J Haematol. 2016;174:767-75.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The spectrum of subclonal TP53 mutations in chronic lymphocytic leukemia: A next generation sequencing retrospective study. Hematol Oncol. 2022;40:962-75.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]







