Translate this page into:
Application of Artificial Neural Networks for Prognostic Modeling in Lung Cancer after Combining Radiomic and Clinical Features
Address for correspondence Irfan Ahmad, DNB, Department of Radiation Oncology, Rajiv Gandhi Cancer Institute and Research Centre, Sector 5, Rohini, New Delhi 110085, India. irfan.a@icloud.com
-
Received: ,
Accepted: ,
This article was originally published by Thieme Medical and Scientific Publishers Private Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective This study was aimed to investigate machine learning (ML) and artificial neural networks (ANNs) in the prognostic modeling of lung cancer, utilizing high-dimensional data.
Materials and Methods A computed tomography (CT) dataset of inoperable nonsmall cell lung carcinoma (NSCLC) patients with embedded tumor segmentation and survival status, comprising 422 patients, was selected. Radiomic data extraction was performed on Computation Environment for Radiation Research (CERR). The survival probability was first determined based on clinical features only and then unsupervised ML methods. Supervised ANN modeling was performed by direct and hybrid modeling which were subsequently compared. Statistical significance was set at <0.05.
Results Survival analyses based on clinical features alone were not significant, except for gender. ML clustering performed on unselected radiomic and clinical data demonstrated a significant difference in survival (two-step cluster, median overall survival [ mOS]: 30.3 vs. 17.2 m; p = 0.03; K-means cluster, mOS: 21.1 vs. 7.3 m; p < 0.001). Direct ANN modeling yielded a better overall model accuracy utilizing multilayer perceptron (MLP) than radial basis function (RBF; 79.2 vs. 61.4%, respectively). Hybrid modeling with MLP (after feature selection with ML) resulted in an overall model accuracy of 80%. There was no difference in model accuracy after direct and hybrid modeling (p = 0.164).
Conclusion Our preliminary study supports the application of ANN in predicting outcomes based on radiomic and clinical data.
Keywords
artificial intelligence
artificial neural network
machine learning
nonsmall cell lung cancer
prognostic modeling
radiomics
Introduction
There has been an exponential increase in data extracted from clinical trials in lung cancer, especially nonsmall cell lung carcinoma (NSCLC).1 Despite an abundance of clinical data, prognostic models based on conventional patient, tumor, and treatment-related parameters often do not explain the variance in survival outcomes, largely due to complex interactions between these factors, as well as intratumoral heterogeneity.2 An elegant method of analyzing intratumoral heterogeneity is to extract radiomic features (quantitative imaging biomarkers) from imaging data and correlate them with outcomes.3 Due to high-dimensional and nonparametric nature of radiomic features, outcome prediction could be enhanced by semiautomated analyses utilizing machine learning (ML) and artificial intelligence (AI) techniques.
The objectives of this preliminary study were to (1) assess the feasibility of combining high-dimensional radiomic data with minimal clinical data in predicting treatment outcome and (2) compare the prognostic accuracy of AI modeling methodologies.
Materials and Methods
Patient Population
A publicly available anonymized computed tomography (CT) dataset of NSCLC patients with embedded tumor segmentation and survival data (NSCLC-radiomics), composed of 422 patients, was selected from The Cancer Imaging Archive (TCIA).3,4,5 The demographic details of the analyzed dataset are available in open access format and therefore will not be repeated here, except those pertaining to our analysis.3,4
All patients had inoperable and histologically confirmed NSCLC across American Joint Committee on Cancer (AJCC) stages I to IIIB and were treated with either radical radiotherapy alone (n = 196) or concurrent chemoradiation (CCRT; n = 226). Radiotherapy (RT) in both groups was delivered with individualized dose-escalation and twice-daily treatment (59.4–79.2 Gy in 1.8 Gy/Fx delivered twice daily in RT alone group and 61–69 Gy in 1.5 Gy/Fx delivered twice daily along with carboplatin and gemcitabine in CCRT group).
All patients underwent 18FDG (flourodeoxyglucose) PET-CT (positron emission tomography–computed tomography) scan for RT treatment planning (Siemens Somatom Sensation 16 with an Ecal Accel PET scanner; Siemens Healthineers, Erlangen, Bavaria, Germany) with a standardized radiotracer injection and image acquisition protocol. A spiral CT with a slice thickness of 3 mm with intravenous contrast was performed covering the complete thoracic region. Gross tumor volume (GTV) segmentation was based on fused PET-CT images with fixed window level settings of both CT (lung W 1,700; L–300, mediastinum W 600; L 40) and PET scan (W 30,000; L 15,000).
The total number of clinical features provided with the dataset for every patient was eight (age, gender, cT, cN, cM, stage, histology, and survival time). The entire dataset was reviewed for completeness of clinical data and tumor segmentation by the authors. A total of 119 patients were excluded from this analysis due to the unavailability of segmentation and/or missing outcome data and/or incorrect segmentation.
Radiomics Feature Extraction
The CT datasets of 303 patients (in DICOM [digital imaging and communications in medicine]-RT format) selected for analysis underwent the following sequence for feature extraction: (1) image preprocessing and standardization, (2) image import into Computation Environment for Radiation Research (CERR) on Matlab (Mathworks, Massachusetts, United States), (3) automated predefined three-dimensional radiomics feature extraction on Matlab, and (4) automated feature export into an analyzable database.6 The workflow was executed by running a custom batch extraction script written in Matlab. A total of 123 radiomics features (without wavelet transformation) were extracted from each patient after noise reduction and three features were excluded due to redundancy. All extracted features were compliant with the Imaging Biomarker Standardization Initiative.7
Statistical Analysis
First, univariate and multivariate analyses were performed to determine the survival probability based on clinical features alone. Next, unsupervised ML analyses were performed by combining clinical and radiomic features, which served as an input for clustering techniques (two-step cluster and K-means cluster). Subsequently, supervised direct AI modeling was performed using artificial neural networks (ANNs; radial basis function network and multilayer perceptron network) after splitting the entire dataset into training and validation cohorts, with clinical and radiomic features serving as the input layer and clinical outcome (alive/dead) as the binary output layer. The prognostic accuracy of the generated model was assessed by a receiver operating characteristic (ROC) curve analysis. Finally, hybrid AI modeling was performed in which ML was used to assess the most important predictors of differential outcome. Predictors with a greater than 50% normalized importance were identified and served as the input layer for AI analysis (with multilayer perceptron network) with clinical outcome (alive/dead) as the output layer. The prognostic accuracy of the generated model was subsequently assessed by a ROC curve analysis. Direct and hybrid AI modeling results were compared on the basis of ROC analysis predicated on the assumption that the ANN algorithms iteratively sampled different sets of patients, resulting in a predictor population distribution for each model being independent of each other.8
Patient-related characteristics and outcomes were imported from the database into IBM SPSS version 23 (Armonk, New York, United States) and survival statistics were generated using the Kaplan–Meir method. Uniform overall survival (OS) estimates were calculated for different variables and differences compared using two-sided log-rank (Mantel–Cox) method and the two-sided p < 0.05 was considered statistically significant. For comparison of ROC curves, one-sided p < 0.05 was considered statistically significant.
The entire schema and results of the study are shown in Fig. 1.
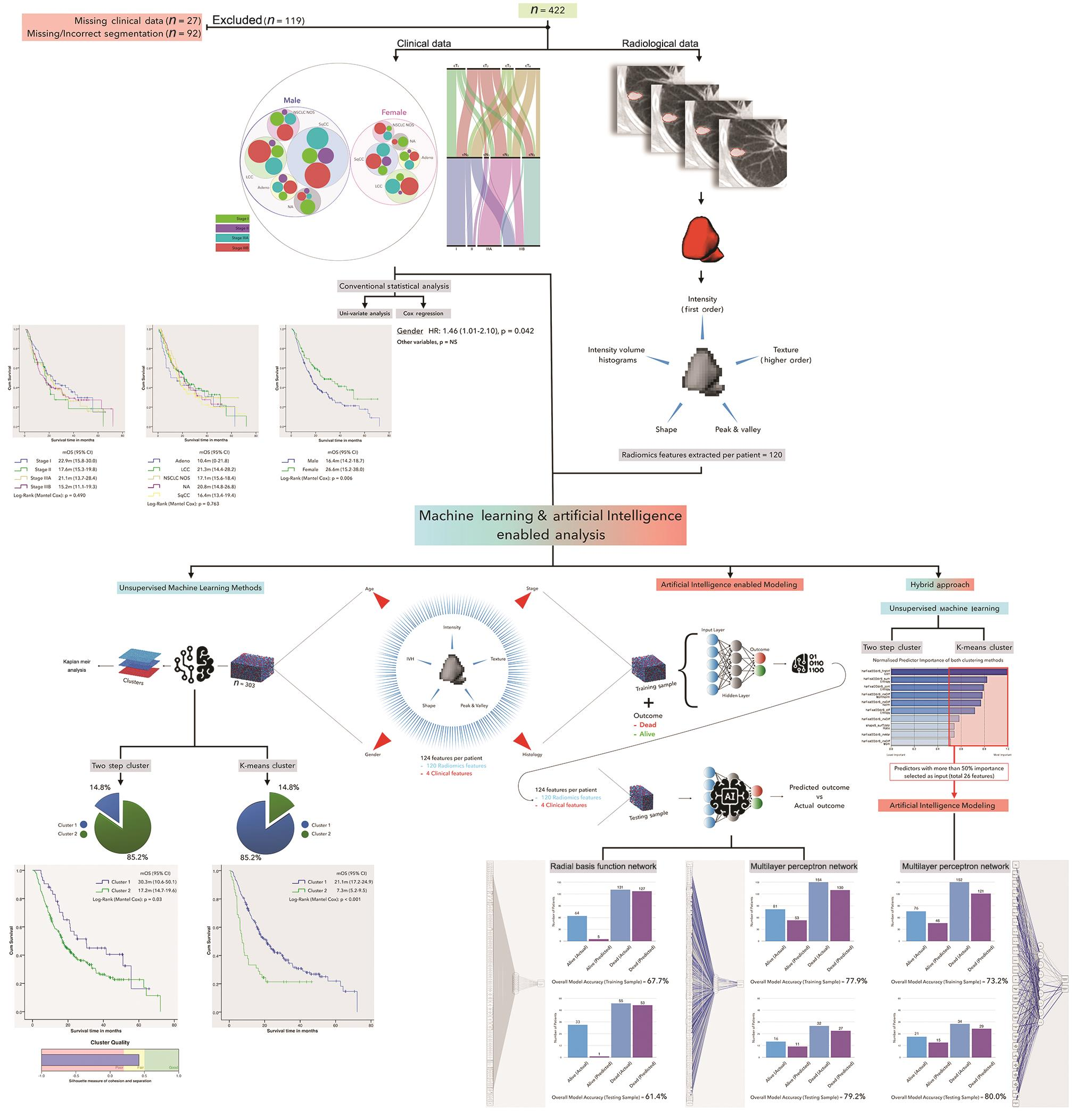
-
Fig. 1 Summary of study schema and results. CI, confidence interval; HR, hazard ratio; IVH, intensity volume histograms; LCC, large cell carcinoma; mOS, median overall survival; NA, not available; NOS, not otherwise specified; NS, not significant; NSCLC, nonsmall cell lung carcinoma.
Results
The results of univariate and multivariate analyses performed based on clinical features only were not significant, with the exception of gender (log-rank [Mantel–Cox] p = 0.006; hazard ratio = 1.46 [range: 1.01–2.10], p = 0.042). The results of the unsupervised ML methods separated the entire cohort into two cohorts with distinctly different prognoses. Utilizing the two-step cluster method segregated the cohort into two clusters, with a median survival of 30.3 months (95% confidence interval [CI] = 10.6–50.1 months) and 17.2 months (95% CI = 14.7–19.6 months), respectively (log-rank [Mantel–Cox] p = 0.03). The K-means clustering method also segregated the cohort into two clusters, with a median survival of 21.1 months (95% CI = 10.6–50.1 months) and 7.3 months (95% CI = 14.7–19.6 months), respectively (log-rank [Mantel–Cox] p < 0.001]. Furthermore, an increase in the number of clusters did not improve the quality of clustering for both methods (results not shown).
The results of direct ANN modeling with radial basis function network (utilizing a single hidden layer) resulted in an overall model accuracy of 67.7% in predicting the primary outcome for the training dataset, which comprised 70% of the entire dataset. However, the accuracy decreased to 61.4% when it was applied to the validation dataset, composed of 30% of the entire dataset. ANN modeling with multilayer perceptron network (utilizing a single hidden layer) resulted in an overall model accuracy of 77.9% in predicting the primary outcome for the training dataset, which comprised 83% of the entire dataset. The accuracy increased to 79.2% when it was applied to the validation dataset, composed of 17% of the entire dataset. On applying a ROC analysis on this model, the area under the curve (AUC) was 0.87. The accuracy of both models did not improve further by adding additional hidden layers or by altering the proportion of patients in the training and validation datasets (results not shown).
Finally, a hybrid modeling approach was performed, in which the predictors with greater than 50% normalized importance discovered on clustering (with both K-means and two-step cluster) served as the input for ANN modeling by multilayer perceptron network. This reduced the number of inputs to 26 features and the results of ANN modeling revealed an overall model accuracy of 73.2% in predicting the primary outcome for the training dataset, which comprised 80% of the entire dataset. The accuracy increased to 80% when it was applied to the validation dataset, composed of 20% of the entire dataset. On applying a ROC analysis on this model, the AUC was 0.84. The accuracy of this approach did not improve further by adding additional hidden layers, increasing/decreasing the predictor importance cut-off, or by altering the proportion of patients in the training and validation datasets (results not shown).
One-tailed comparison of ROC curves generated by the direct ANN modeling and hybrid modeling approach did not reveal a statistically significant difference (Fig. 2; p = 0.164).
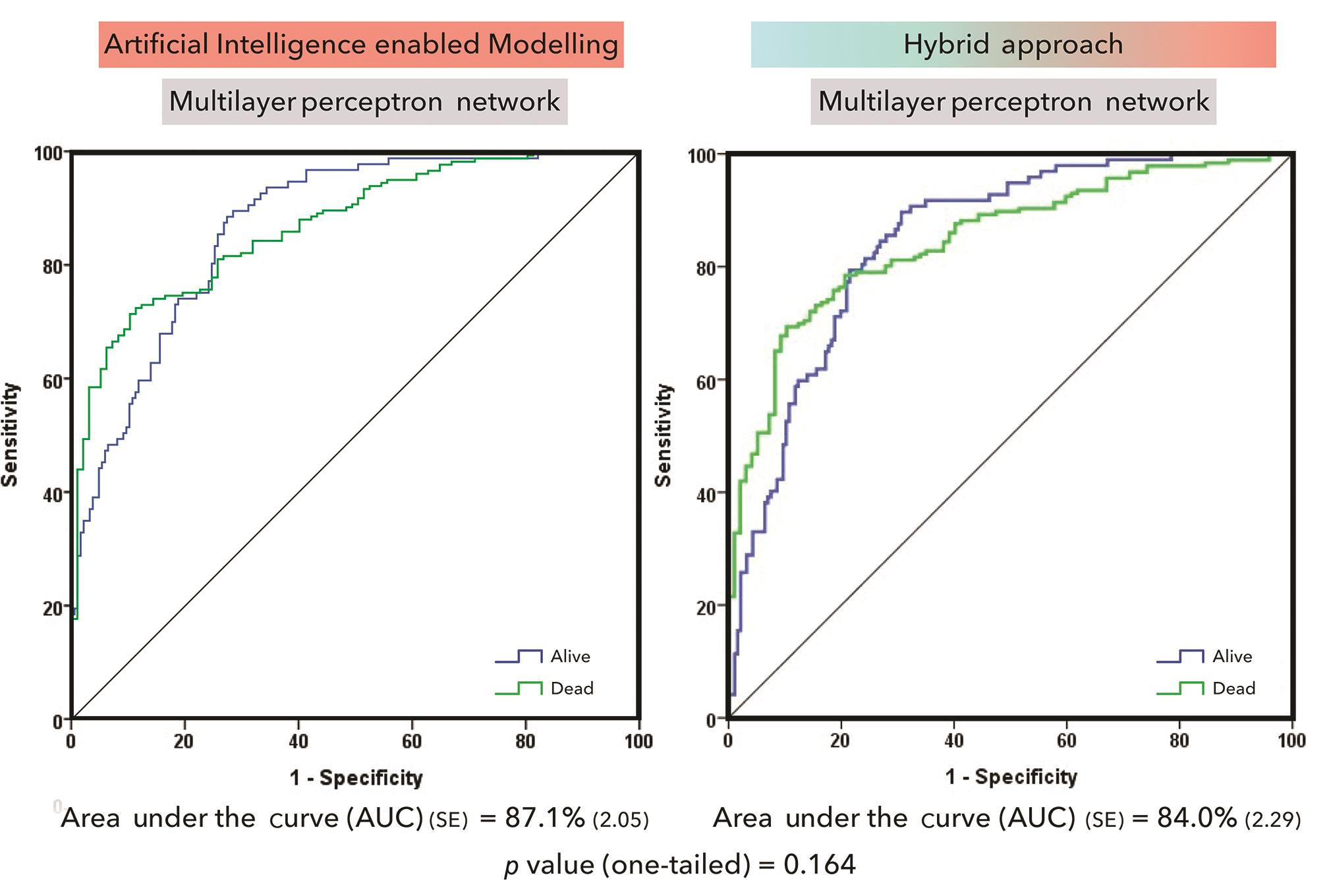
-
Fig. 2 Comparison of direct and hybrid artificial neural network modeling.
Discussion
With an increasing incidence of patients with advanced NSCLC, usually not amenable to surgery, the data required to generate prognostic models for risk stratification and treatment intensification has grown exponentially. A multitude of clinical factors, molecular markers, and gene signatures have been explored, yet a correlation between the above mentioned and many other possibly unknown factors has not been established and patients continue to exhibit varied survival outcomes within a given stage.2 In this study, we investigated the ability of radiomic features extracted from the tumor combined with basic clinical data in predicting the probability of an adverse outcome, utilizing ML and AI techniques.
We found that clinical features only were not predictive of differences in outcome, except for gender and this could be attributed to the limited number of variables analyzed. The decision to include a limited number of clinical variables in our analysis was by design, as we hypothesized that intratumoral radiomic features could predict our primary outcome with greater certainty. The second part of our analysis demonstrated that upon utilizing unsupervised clustering techniques, there were indeed radiomic features which were able to sort the patient cohort into clusters with remarkably different survival outcomes. We also demonstrated that ANN’s could be trained to recognize intratumoral radiomic features and their interdependencies to predict outcomes with up to 79.2% accuracy.
Our results also challenge the methodology adopted by other investigators exploring the application of radiomic data in the prediction of survival outcomes in NSCLC.3,9,10 Due to the high-dimensional nature of extracted radiomic features, performing conventional statistical modeling requires selection of the most informative features. This could potentially lead to selection bias and ignore other less informative features.11 We hypothesized that combining even relatively less informative features, would in concert lead to equivalent/stronger prediction than that obtained from selected features. Our results demonstrated that utilizing a hybrid approach to ANN modeling did not improve the performance in comparison to direct ANN modeling.
Criticism of our analysis may arise when considering that this is at present an internally validated algorithm. Our research group will attempt to externally validate this algorithm on another publicly available dataset and our institutional dataset soon. Furthermore, the exclusion of 119 patients from the original dataset due to missing data/segmentation could have reduced the accuracy of our model. We deliberately chose to not introduce our interobserver bias by segmenting those patients whose CT datasets did not have tumor segmentations. A future avenue of analysis could also be to study the influence of interobserver variation in tumor segmentation on the accuracy of our prediction algorithm, all other factors being the same. Finally, the weaknesses associated with retrospective analyses are also applicable to this study.
Conclusion
Our analysis provides a proof of concept on the application of ML- and AI-based modeling in predicting patient outcomes utilizing a combination of radiomic features and clinical data.
Conflict of Interest
None declared.
References
- Multimodality treatment of advanced non-small cell lung cancer: where are we with the evidence? Curr Surg Rep. 2018;6(02):5.
- [Google Scholar]
- The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(01):39-51.
- [Google Scholar]
- Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006.
- [Google Scholar]
- The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(06):1045-1057.
- [Google Scholar]
- Technical Note: Extension of CERR for computational radiomics: a comprehensive MATLAB platform for reproducible radiomics research. Med Phys. 2018;45:3713-3720. 10.1002/mp.13046
- [Google Scholar]
- Zwanenburg A, Leger S, Valli’eres M, L ̈ock S. Image biomarker standardisation initiative. Available at: https://arxiv.org/abs/1612.07003 Accessed November 15, 2019
- A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148(03):839-843.
- [Google Scholar]
- Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281(03):947-957.
- [Google Scholar]
- Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2014;90(04):834-842.
- [Google Scholar]
- Vulnerabilities of radiomic signature development: the need for safeguards. Radiother Oncol. 2019;130:2-9.
- [Google Scholar]







